Educating an Intelligent Design Creationist: Introduction
Educating an Intelligent Design Creationist: Pervasive Transcription
Educating an Intelligent Design Creationist: Rare Transcripts
The Specificity of DNA Binding Proteins
It is absolutely essential that you understand the basic biochemistry of DNA binding proteins if you want to interpret the ENCODE results and the controversy surrounding junk DNA. You might think this is a given since almost everyone involved in the discussion has had some exposure to biochemistry in undergraduate courses. Unfortunately, most of these courses don't teach that stuff anymore1 so we've raised a generation of scientists who were never exposed to the facts.
THEME:
Transcription
I've blogged about this many times in the past. The model system is the lac repressor since so much work has been done on DNA binding over the past 40 years [DNA Binding Proteins] [Repression of the lac Operon]. We know a great deal about the thermodynamics and kinetics of binding of lac repressor to DNA. It binds specifically to three DNA sequences (operators) near the promoter of the lac operon. The three operators have slightly different sequences and this affects the strength of binding. It was results like this that led to the concept of a "consensus sequence"—a DNA sequence that represents the ideal binding site. Regions of DNA that resemble the consensus sequence will be bound less tightly. There's a progression in strength of binding that ranges from very strong binding to the consensus sequences all the way down to very weak binding to a DNA sequence that has no resemblance to the consensus.
Why do specific DNA binding proteins also bind to random sequences of DNA? There are two good reasons. First, it is impossible for DNA binding proteins to discriminate absolutely between sequences that resemble the binding site and those that don't. All DNA binding proteins have to recognize the sugar-phosphate backbone of DNA before they can probe the exact sequence of the stacked bases in the interior of the molecule.

The second reason is more important. Here's how I described it in: DNA Binding Proteins.
Now, here's the important point: all specific DNA binding proteins also bind DNA non-specifically. In many cases it's part of the search mechanism for the specific binding site. In the case of lac repressor, for example, the protein binds to any old place on the DNA molecule and slides along the DNA searching for a specific binding sequence. After sliding for a second or so it falls off and re-binds to another part of the DNA molecule.You'll have to read that post for more details. Just keep in mind that all specific DNA binding proteins must also bind non-specifically.
The lac repressor is the very best protein at discriminating between specific and non-specific sites. The non-specific binding constant is Ka ~ 106 M-1. That may not mean very much to most of you but it's pretty high. It means that lac repressor binds pretty tightly to any old sequence of DNA. This value is about the same for all DNA binding proteins, including RNA polymerase. The specific binding constant (equilibrium association constant) represents the strength of binding to the ideal operator site (the consensus sequence). It's value is Ka ~ 1013 M-1. That's seven orders of magnitude stronger than non-specific binding. No other DNA binding protein binds so strongly to its target site.
In spite of this huge difference, most lac repressor molecules inside an E. coli cell will be sitting on sites other than the operators at any one time. That's because there are 4.6 million non-specific binding sites and only three specific binding sites. If the E. coli genome was full of extra DNA, like the mammalian genome, then there would be 6.4 billion binding sites. That's why eukaryotic cells need so many more molecules of each transcription factor compared to bacteria—most of them are sitting where they're not supposed to be (Yamamoto and Alberts, 1976).
 RNA polymerase binds specifically to promoter sequences. It adopts the same binding mechanism as other specific DNA binding proteins; namely, it binds non-specifically then slides along DNA until it finds a promoter sequence. The sequence of a eukaryotic promoter is not very well defined so in most cases eukaryotic RNA polymerase needs help in the form of a nearby transcription factor to bring it to the correct transcription initiation site.
RNA polymerase binds specifically to promoter sequences. It adopts the same binding mechanism as other specific DNA binding proteins; namely, it binds non-specifically then slides along DNA until it finds a promoter sequence. The sequence of a eukaryotic promoter is not very well defined so in most cases eukaryotic RNA polymerase needs help in the form of a nearby transcription factor to bring it to the correct transcription initiation site. Nevertheless, the basic concept is the same. We know the kinetics and binding constants for E. coli RNA polymerase and they produce the expected distribution [see How RNA Polymerase Binds to DNA for all the references]. A significant percentage of RNA polymerase molecules are sitting at sites other than genes and promoters. The situation is much worse in mammals with large genomes. You need tens of thousands of RNA polymerase molecules in order to ensure that promoters will be occupied. Most of these are sitting at sites that fortuitously resemble real promoters or they are bound to a transcription factor that is also at a non-specific site.
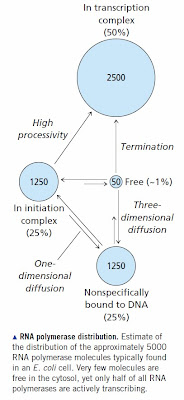 None of this is controversial once you have read the papers and understand the principles of DNA binding. It is straightforward biochemistry/molecular biology at the undergraduate level. We expect the mammalian genome to be covered with non-functional transcription factor binding sites and bound RNA polymerase molecules. Many of these will be in pre-initiation complexes and many will actually be the sites of spurious transcription by accident.
None of this is controversial once you have read the papers and understand the principles of DNA binding. It is straightforward biochemistry/molecular biology at the undergraduate level. We expect the mammalian genome to be covered with non-functional transcription factor binding sites and bound RNA polymerase molecules. Many of these will be in pre-initiation complexes and many will actually be the sites of spurious transcription by accident. This is exactly what Kevin Stuhl (2007) was talking about when the preliminary ENCODE results were published six years ago. He said ...
The issue of transcriptional noise has become increasingly important, because recent studies in a wide range of eukaryotic organisms indicate that there is far more transcription than expected from the classical view of the transcriptome. Here, on the basis of experimental observations, including a recent analysis of genome-wide distribution of Pol II [RNA polymerase II], I estimate that only 10% of the elongating Pol II molecules in the yeast Saccharomyces cerevisiae are engaged in transcription that initiates from conventional promoters and that the remaining 90% of the elongating Pol II molecules represent transcriptional noise. Furthermore, these calculations suggest that the specificity of Pol II initiation (an approx 104-fold difference between an optimal site and an average genomic site) is comparable to that of sequence-specific DNA-binding proteins and other biological processes considered to be specific.The ENCODE preliminary result confirmed back in 2007 what we knew about the properties of transcription factors and RNA polymerase. The completed project extended this result to the entire genome. The ENCODE experiments detected 636,336 sites where there were bound transcription factors (119 different factors) and tens of thousands of sites where RNA polymerase was bound. It would have been surprising if these sites had NOT been found.
Andyjones asks,
And how do we know that RNA polymerase is meant to bind only promoters? Is it safe to assume that when RNA polymerase binds to other sites, this must be accidental or unintentional? Could it be that these other sites are meant to be transcribed only rarely? I don’t know, but I would like to know. Why don’t we encourage scientists to take a deeper look? Oh great, that’s what ENCODE are doing.I hope I've answered your question. We have excellent reasons for believing that many of those bound RNA polymerases are sitting at spurious binding sites that aren't real promoters. That doesn't mean that all of them are at non-functional sites but it does mean that most of them have to be at non-promoters or else our understanding of the basic biochemistry of binding is seriously flawed.
 Andyjones continues ...
Andyjones continues ...So, Larry thinks that rare transcription (which he believes is due to RNAP binding sites that are not recognised promoters) indicates accidental transcription. That is his argument. Ironically, Larry is using a design heuristic here (assuming that promoter means ‘bind only here’). All I am suggesting (not claiming to know for sure) is that perhaps the correct design heuristic should be that a promoter really means ‘bind more often here’? If so, there would be no reason to assume non-function.I hope that andyjones will continue this conversation if there's still something he doesn't understand about the properties of DNA binding proteins. I recommend that he read an introductory biochemistry/molecular biology textbook if he's still confused. I know of one that I'd recommend.
It is quite reasonable now to expect that details of actual function will subsequently be found for much of the genome. Therefore we should keep looking for that function.

1. It's not covered on the MCAT!
[Image Credit: Moran, L.A., Horton, H.R., Scrimgeour, K.G., and Perry, M.D. (2012) Principles of Biochemistry 5th ed., Pearson Education Inc. [Pearson: Principles of Biochemistry 5/E] © 2012 Pearson Education Inc.]
Struhl, K. (2007) Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nature Structural & Molecular Biology 14:103-105. [doi: 10.1038/nsmb0207-103]
Yamamoto, K.R. and Alberts, B.M. (1976) Steroid Receptors: Elements for Modulation of Eukaryotic Transcription. Ann. Rev. Biochm. 45:721-746. [

18 comments :
Thanks for these wonderful posts L. You're really going out of your way to explain all this considering that many IDers have already said that even if much of the genome was proven to be junk it still wouldn't be a problem for ID. I guess wars are won by taking one hill at a time.
We have excellent reasons for believing that many of those bound RNA polymerases are sitting at spurious binding sites that aren't real promoters.
An obvious way of testing this would be technical replication. If ENCODE do the same ChIP experiment for the same transcription factor in the same tissue type twice (or more), do they find the same binding sites each time? Did they try this?
Spurious binding sites can be replicated. It is the sites that are spurious, not the binding. One way to test would be to see how far they are conserved. But then we have the problem of distinguishing those that are new but meaningful, from those that are authentically spurious. Quite the challenge. But the more samples the better. Another way to test is whether they do come together with other transcription factors or DNA-binding proteins so as to make much better sense of such a binding site ... et cetera.
Spurious sites are expected because the probability for something that looks like a binding site over a huge genome is somewhat high.
And how do we know that RNA polymerase is meant to bind only promoters?
One well understood class of such sites are enhancers - when you do ChIP-seq against Pol2, often it will cross-link quite robustly to enhancers because of the looping of the latter to the promoter. And sometimes it might transcribe them though I am personally not at all convinced in the functional importance of the whole eRNA story.
There is no a priori reason why DNA polymerase should have any greater specificity for DNA sequences than RNA polymerase, but the licensing of DNA replication origins is observed to be very strictly controlled so it is NOT the case that spurious biochemical activity is a necessary consequence of the limited site specificity of any single given protein, contrary to the implication in the post. You could rather argue that spurious transcription is of much less consequence than spurious replication, and that therefor transcription doesn't need such stringent licensing and therefore spurious transcription is to be expected, but spurious activity as a necessary consequence of the thermodynamics of protein/DNA binding for higher order protein complexes is false.
How many DNA polymerases exist in a cell at any given time compared to RNA polymerases?
I don't know, but I could speculate that they're normally only upregulated and active around mitosis. So it's not so much that they're much more specific and selective than RNA polymerase, but most of the time are so heavily downregulated that spurious replication of some stretch of DNA almost never takes place?
Note that the initiating event of DNA replication is RNA priming and that the DNA polymerase primase is itself an RNA polymerase.
Well in the end there's more to it than that. The off chance that you'd get spuriuos DNA replication isn't so much because of the "specificity of the DNA polymerase" compared to that of RNA polymerase. It's because in general, DNA replication requires that whole ridiculous origin-recognition complex and so on before you even get that RNA primer made.
I guess what you're doing is throwing that whole thing under the umbrella of "specificity" of DNA polymerase?
Correct. Using higher-order protein complexes it is possible to very strictly control the biochemical activity of DNA binding proteins. That level of control might not be happening with transcription, but it's not because that level of control is not possible. In other words you can't successfully argue that the multitude of transcripts detected by ENCODE are necessarily spurious due to the lack of specificity of individual DNA binding proteins.
Alright thank you for the clarification.
whimples says,
... spurious activity as a necessary consequence of the thermodynamics of protein/DNA binding for higher order protein complexes is false.
Most of the "activity" that ENCODE measured is just binding and nonspecific binding is a necessary consequence of the properties of specific DNA binding proteins.
What you mean is that nonspecific binding of RNA polymerase holoenzyme doesn't always lead to spurious transcription. This is true but sometimes it does, especially when assisted by nonspecifc binding of nearby activators. RNA polymerase holoenzyme is fully capable of initiating polynucleotide synthesis all on its own ... unlike DNA polymerase. DNA polymerases will be bound to DNA all over the genome but DNA replication is still pretty much confined to origins.
I maintain that spurious transcription is bound to happen, given what we know about RNA polymerase. That's why we use calf thymus DNA in the assay for E. coli RNA polymerase activity in our undergraduate lab course.
That's why we use calf thymus DNA in the assay for E. coli RNA polymerase activity in our undergraduate lab course.
If that was not enough for an "aha!" moment, I don't know what would be.
I don't think you've read the papers - reproducibility was a core component of all experimental design and analysis that was done. Everything except for DGF was done in replicates.
You may want to start with this one, then read all the biology-focused papers:
Li Q., J. Brown, H. Huang, and P. Bickel, 2011 Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5:1752–1779.
One clarification I think is needed here - when you do RNA-seq, you can capture spurious transcription, because even one molecule of RNA can end up in your libraries and if you sequence deep enough, you will eventually start seeing all the very rare transcripts.
With ChIP-seq it's a very different situation because you require a large number of distinct fragments to be sequenced around a binding site. As a consequence you can not identify non-specific binding from ChIP-seq, because that would by definition give you only one sequencing read, and that is just background that nobody pays any attention to. The sites that are identified are real sites, and when you apply IDR or other reproducibility criteria, you are asking for them to be consistently and robustly present in two different experiments. So proteins do bind to those sites and they do so quite specifically, and this is very different from random non-specific association with DNA. Not only that, but that model only really works in such a straightforward way in prokaryotes - in eukaryotes you have chromatin which depending on its local state can prevent most transcription factors from binding to DNA quite effectively.
Now, whether each of those binding sites has a function in regulating transcription is a different question (and the answer is IMO highly unlikely to be positive for all of them)
And I guess the existence of transcription does not in itself invalidate the fact that the transcribed DNA may be junk - it simply generates junk RNA. Arguments against junk are often made on the basis of 'cost' which is assumed rather than demonstrated. Junk DNA costs something, turning it into junk RNA a bit more, but they roll up into an overall per-base-pair cost: two molecules of dNTP per cell cycle (locked) plus however many molecules of NTP are polymerised off it (NMP recycled).
Georgi Marinov says,
So proteins do bind to those sites and they do so quite specifically, and this is very different from random non-specific association with DNA.
Nonspecific binding ranges all the way from fairly weak binding to random DNA sequences to fairly strong binding to nonfunctional sites that closely resemble the specific functional binding site. Since most specific binding sites (consensus sequence) are ten base pairs or less, it is statistically certain that many similar sites will occur in a large genome full of junk. These will be readily detected as reproducible binding sites by a variety of different assays.
Many of these nonspecific sites will be masked by being located in "closed" chromatin regions just as truly functional sites are masked. The ones that are accessible will tend to be in "open" regions near where genes are being transcribed. This results in some tissue specificity but that doesn't mean that the sites are functional.
According to my definition "specific binding" refers to binding that is driven by sequence composition and sequence binding specificity (this could be secondary, through a protein-protein interaction), happens sufficiently frequently for it to be cross-linked, identified by a ChIP-seq peak caller and be reproducible across replicates.
"Functionality" is a largely orthogonal concept to that.
It is possible that we differ in the exact meaning we put in the terms when we use them.
LM: What you mean is that nonspecific binding of RNA polymerase holoenzyme doesn't always lead to spurious transcription.
No. What I mean is that strictly controlled biochemical activity of DNA binding proteins in a human-sized genome is possible, as shown by DNA replication. Your post seemed to be asserting that it was not possible to strictly regulate the activity of DNA binding proteins, which is obviously not the case. For what it's worth, I agree with you that most the ENCODE detected transcripts are spurious and without function.
Post a Comment